Secondary Structure Predictions

Protein Secondary Structure Prediction Youtube Other sites for secondary structure predictions include: jpred4 is the latest version of the popular jpred protein secondary structure prediction server which provides predictions by the jnet algorithm, one of the most accurate methods for secondary structure prediction. in addition to protein secondary structure, jpred also makes predictions. A limit of 200 sequences are allowed per batch submission. you may submit several batch jobs, but there is a hard limit of 4,000 sequence predictions in total per user per day. please let us if this limit is a problem for you and we'll look at temporarily changing it. by default jpred searches the pdb before doing a prediction.
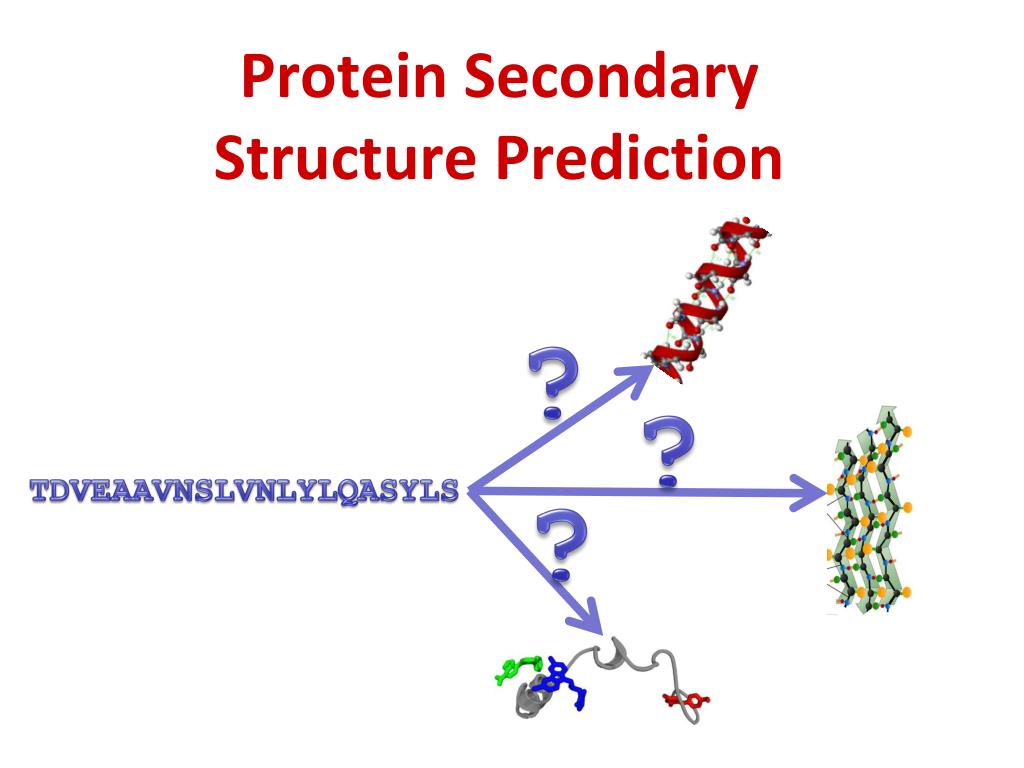
Ppt Protein Secondary Structure Prediction Powerpoint Presentation Sequence name. amino acid sequence (using one letter codes) query sequence in the fasta format file. e mail address (optional) sable server can be used for prediction of the protein secondary structure, relative solvent accessibility and trans membrane domains providing state of the art prediction accuracy. Protein secondary structure prediction (pssp) is one of the subsidiary tasks of protein structure prediction and is regarded as an intermediary step in predicting protein tertiary structure. if protein secondary structure can be determined precisely, it helps to predict various structural properties useful for tertiary structure prediction. Prediction algorithm. as with jpred3, jpred4 makes secondary structure and residue solvent accessibility predictions by the jnet algorithm (11,31).however, in jpred4, the jnet 2.0 neural network based predictor has been retrained to make jnet 2.3.1 by 7 fold cross validation using one representative for each of the 1358 scope astral v.2.04 superfamily domain sequences (). Protein secondary structure prediction (pssp) is a fundamental task in protein science and computational biology, and it can be used to understand protein 3 dimensional (3 d) structures, further, to learn their biological functions. in the past decade, a large number of methods have been proposed for pssp.

Comments are closed.