Protein Prediction I Beginners Lecture 9 Secondary Structure

Protein Secondary Structure Prediction Youtube Types of protein structure predictions • prediction in 1d –secondary structure –solvent accessibility (which residues are exposed to water, which are buried) –transmembrane helices (which residues span membranes) • prediction in 2d –inter residue strand contacts • prediction in 3d –homology modeling –fold recognition (e.g. via. Protein secondary structure prediction. empirical (statistical) approachesextrapolating the statistics from known structures: ala, gln, leu and met are found in α helices. pro, gly, tyr and ser usually in α helices. (bulky. pro is found to be a ring prevents n n 4 h bonds formation) 9. ‘helix breaker’.

Protein Secondary Structure Prediction Youtube Uses a large database of 3 residue and 9 residue fragments, taken from structures in the pdb. monte carlo sampling algorithm proceeds as follows: start with the protein in an extended conformation. randomly select a 3 residue or 9 residue section. find a fragment in the library whose sequence resembles it. Most of what happens over evolution is small changes that tune the function of the protein. it’s highly unlikely to have a small change that alters the folded structure. during evolution, sequence changes more quickly than structure. also, there only appear to be 1,000–10,000 naturally occurring protein folds. See the results for secondary structure prediction for one protein. in this example, the average propensity for four contiguous amino acids is calculated (starting with amino acids 1 4, then amino acids 5 8, etc, and continuing to the end of the polypeptide). next this process is repeated for contiguous stretches 2 5, 6 9, etc, and continuing. Proteus2 is a web server designed to support comprehensive protein structure prediction and structure based annotation. proteus2 accepts either single sequences (for directed studies) or multiple sequences (for whole proteome annotation) and predicts the secondary and, if possible, tertiary structure of the query protein (s).

Protein Prediction I Beginners Lecture 9 Secondary Structure See the results for secondary structure prediction for one protein. in this example, the average propensity for four contiguous amino acids is calculated (starting with amino acids 1 4, then amino acids 5 8, etc, and continuing to the end of the polypeptide). next this process is repeated for contiguous stretches 2 5, 6 9, etc, and continuing. Proteus2 is a web server designed to support comprehensive protein structure prediction and structure based annotation. proteus2 accepts either single sequences (for directed studies) or multiple sequences (for whole proteome annotation) and predicts the secondary and, if possible, tertiary structure of the query protein (s). The most successful methods for predicting secondary structure are based on neural nets. the overall idea is that neural nets are trained to be able to recognize amino acid patterns in known secondary structure units (e.g., helices), and to use these patterns to distinguish between the di erent types of secondary structures. before considering. Lecture 20: secondary structure prediction. lecturer: pankaj k. agarwal scribe: jeffrey j. headd. . with the rapid development of genome sequencing technology, what once took decades can be done in a matter of months. as genomic data continues to be collected at a breakneck pace, interpretation of the results is quickly becoming a daunting effort.
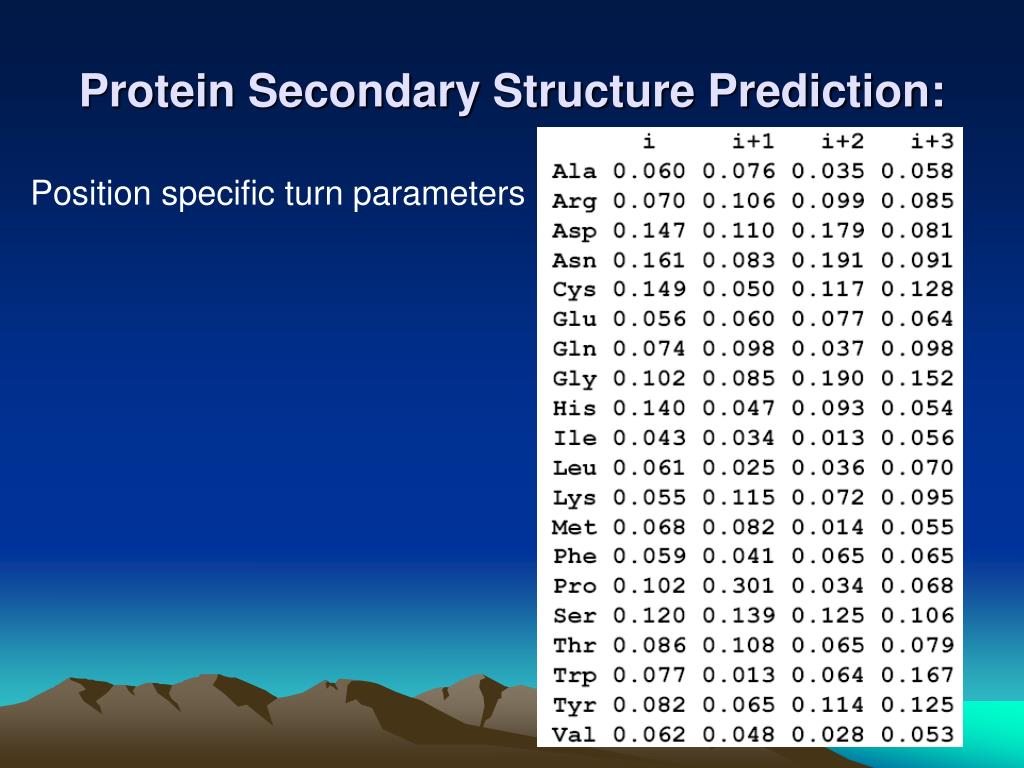
Ppt Protein Structural Organization Powerpoint Presentation Free The most successful methods for predicting secondary structure are based on neural nets. the overall idea is that neural nets are trained to be able to recognize amino acid patterns in known secondary structure units (e.g., helices), and to use these patterns to distinguish between the di erent types of secondary structures. before considering. Lecture 20: secondary structure prediction. lecturer: pankaj k. agarwal scribe: jeffrey j. headd. . with the rapid development of genome sequencing technology, what once took decades can be done in a matter of months. as genomic data continues to be collected at a breakneck pace, interpretation of the results is quickly becoming a daunting effort.
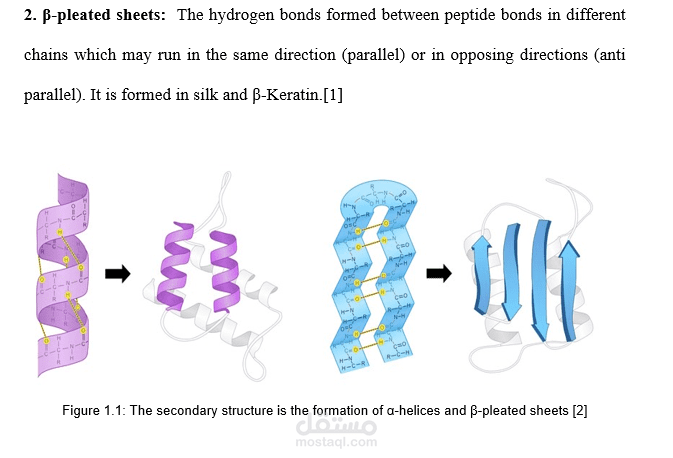
Protein Secondary Structure Prediction щ шішєщ щ

Comments are closed.