Long Read Sequencing Repetitive Regions Youtube

Hudsonalpha Researchers Use Highly Accurate Long Read Sequencing Repetitive regions can be a pain to sequence with traditional techniques. in this video, we explain why oxford nanopore technologies' approach to long read s. Long read sequencing enables us to tackle the challenges posed by repetitive elements, pseudogenes, and other complex regions of the genome in the genetic di.
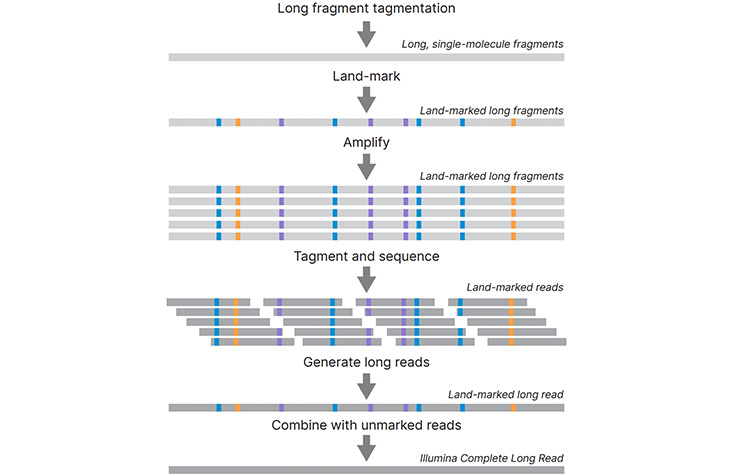
Long Read Sequencing Technology For Challenging Genomes Winnowmap2 enables better long read mapping and more accurate variant calling in repetitive regions of the genome. here we mapped three publicly available hg002 long read sequencing datasets. Long read sequencing, he says, delivers ways to study repetitive and complex genomic regions such as centromeric regions, long repeats and complex structural variants. As demonstrated by their t2t work, long read data shines light on many previously dark regions of the genome, such as telomeres and other highly repetitive regions and complex structural variations. Abstract. about 5 10% of the human genome remains inaccessible due to the presence of repetitive sequences such as segmental duplications and tandem repeat arrays. we show that existing long read mappers often yield incorrect alignments and variant calls within long, near identical repeats, as they remain vulnerable to allelic bias. in the.

Genomics In The Long Read Sequencing Era Trends In Genetics As demonstrated by their t2t work, long read data shines light on many previously dark regions of the genome, such as telomeres and other highly repetitive regions and complex structural variations. Abstract. about 5 10% of the human genome remains inaccessible due to the presence of repetitive sequences such as segmental duplications and tandem repeat arrays. we show that existing long read mappers often yield incorrect alignments and variant calls within long, near identical repeats, as they remain vulnerable to allelic bias. in the. Abstract. full length analysis of genes with highly repetitive sequences is challenging in two respects: assembly algorithm and sequencing accuracy. the de bruijn graph often used in short read assembly cannot distinguish adjacent repeat units. on the other hand, the accuracy of long reads is not yet high enough to identify each and every. Now, the emerging long read sequencing (lrs) technologies may offer improvements in the characterization of genetic variation and regions that are difficult to assess with the prevailing ngs approaches. lrs has so far mainly been used to investigate genetic disorders with previously known or strongly suspected disease loci.
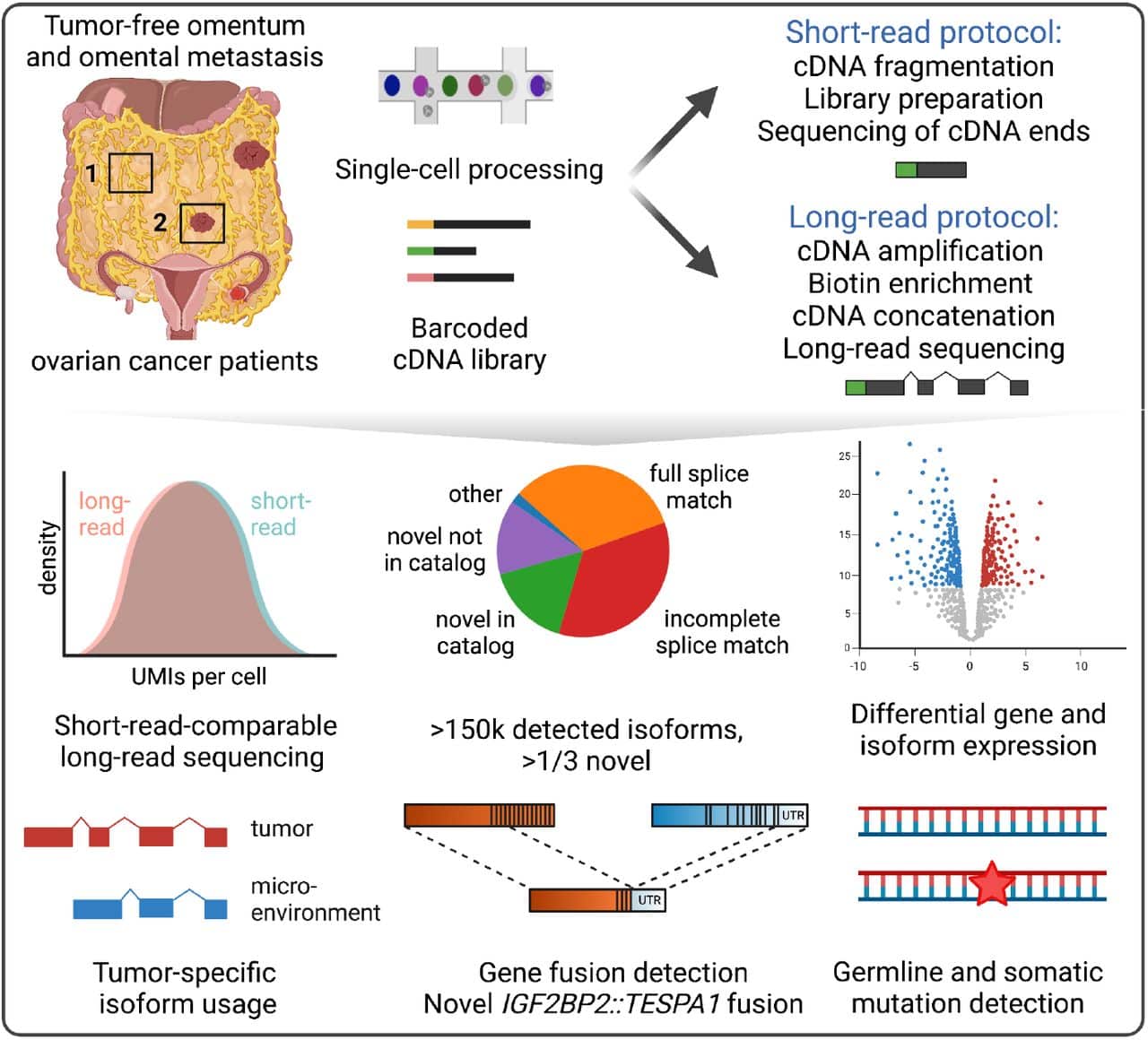
Long Read Sequencing The Key To A More Complete Cancer Transcriptome Abstract. full length analysis of genes with highly repetitive sequences is challenging in two respects: assembly algorithm and sequencing accuracy. the de bruijn graph often used in short read assembly cannot distinguish adjacent repeat units. on the other hand, the accuracy of long reads is not yet high enough to identify each and every. Now, the emerging long read sequencing (lrs) technologies may offer improvements in the characterization of genetic variation and regions that are difficult to assess with the prevailing ngs approaches. lrs has so far mainly been used to investigate genetic disorders with previously known or strongly suspected disease loci.

Comments are closed.